Microbiome
Microorganisms are the silent wheel that functions as the cradle of plant growth, stress signaling, and responses in terrestrial ecosystems. “Microbiome refers to the total genetic material of microbial communities associated with plants in either of these modes and associations”. The main goal of this area of research is to “characterize key microbes that can play a beneficial function in enhancing resistance against environmental stress in a specific ecosystem (aquatic and terrestrial).” We try to understand important questions like:
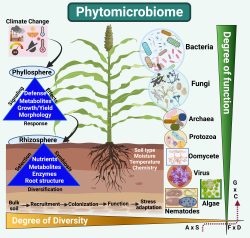
- What are microbial symbionts in the microbiome of keystone halophytes and extremophiles?
- How does core-microbiome function with plants living in extreme conditions and what helps them survive episodes of extreme climate changes?
- How can the core microbiome be isolated to produce stress-competent microbes to increase yield and function of productivities (plants, metabolites or enzymes)?
- What are biosynthetic pathways for essential metabolites and nutrients during plant-microbiome-stress interaction?
- How does the unhealthy plant microbiome can influence the human gut microbiome?
The plant and the soil-associated microbiome has been coined as “second genome” that is highly variable in diversity, abundance and composition. Microbiome diversity and abundances in food crops alternatively influence the human gut microbiome. Healthy soil and crop microbiome is key to healthy human growth. Hence, Khan’s lab is focused on understanding the function of plant microbiome.
Approach:
- Samples collections from different environments (extreme, natural habitats, agriculture)
- Large-scale DNA extractions from soil, plant, microbes
- Sequencing the amplicon 16S/ITS gene using MiSeq Illumina
- Sequencing the metagenome of microbial DNA using NovaSeq 6000 Illumina
- Bioinformatic analysis with the help of UH cluster and available computational resources in the lab
Current projects:
- Environmental Stress Microbiomes: We are currently working on to understand the complexity of rhizospheric microbiome diversity and function during episodes of flooding, drought and temperature across tolerant and susceptible genotypes. This work is in progress with collaboration of University of Missouri.