Molecular Stress Physiology
By understanding the functions and mechanisms in morphology, biochemical, metabolites, and molecular levels and their responses to different environmental changes, one can select specific traits and abilities to develop improved plant and microbial functions and benefits. Stress physiology includes biochemistry, functional biotechnology, computational and synthetic plant biology, growth and development, metabolism, transport and translocation, plant-microbe interactions, ecological physiology, and biotic and abiotic stresses. Microbial physiology can reasonably be defined as “structure-function relationships in microorganisms, especially how microbes respond to their environment”.
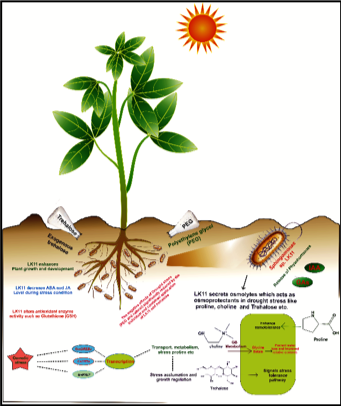
We try to understand important questions like:
- How climatic changes are influencing the molecular signal networks involved in stress tolerance, yield, and growth of plants?
- How does the metabolome change and function in response to environmental stresses?
- What are biosynthetic pathways and gene regulatory dynamics that can be enhanced through gene-editing technologies?
Approaches:
This research work involves several methodologies such as:
- Plant growth in controlled environmental chambers and their exposure to various climatic conditions
- Performing RNA-Seq of leaf, seed, root, shoot, and fruits with the help of Illumina and or nanopore sequencer
- In-depth bioinformatic analysis of RNA-Seq data to understand gene regulatory networks and biosynthetic mechanisms involved during such conditions.
- Performing detailed biochemical, phytohormonal, and antioxidant enzymes assessment with the help of GC/MS SIM, fluorescence spectroscopy, and HPLC
- Metabolomics analysis of targeted and non-targeted ones using LC/MS-MS and Chemo-informatics using available libraries
- Isolating, culturing, and characterization of bioactive microbes from different environments
- Molecular identification by sequencing the amplicon 16S/ITS gene and bioinformatic analysis
- Whole-genome sequencing the bioactive microbial strain with NovaSeq 6000 Illumina and PacBio
- Bioinformatic analysis with the help of UH cluster and available computational resources in the lab
- Identification of gene-clusters and biosynthetic pathways in potential microbes
- Gene cloning, transformation, and genome editing technologies to develop competent microbes
- Extra-Cellular enzymes, biochemical, phytohormonal, and organic acids assessment with the help of GC/MS SIM, fluorescence spectroscopy, and HPLC
- Metabolomics analysis of targeted and non-targeted ones using LC/MS-MS and Chemo-informatics using available libraries